Abstract
Accurate diagnosis of vertebral diseases is vital for prevent ing severe complications, but data imbalance between abundant normal and rare pathological cases poses a substantial challenge to diagnos tic performance. Medical image generation offers a promising solution by synthesizing pathological samples. However, existing diffusion-based methods, pre-trained on natural images, often fall short in capturing complex pathological features due to the pre-training knowledge gap, as well as struggling to obtain precise lesion masks and ensure seam less integration between lesions and the background. To overcome these challenges, we propose a novel diffusion-based medical image genera tion framework called MedSoft-Diffusion, which involves leveraging de tailed medical knowledge to ensure that generated images are not only semantically consistent with the specified pathological conditions but also anatomically accurate. Our framework includes a Medical Semantic Controller (MSC) designed to enhance the alignment between textual prompts and lesion characteristics, ensuring the synthesis of semanti cally accurate pathological images. Furthermore, the Soft Mask Inpaint ing Strategy (SMIS) is proposed to combine soft masks with blurring techniques to improve the realism of synthesized images. Experimental results on two vertebral disease datasets demonstrate notable improve ments in both image quality and classification performance using our approach.
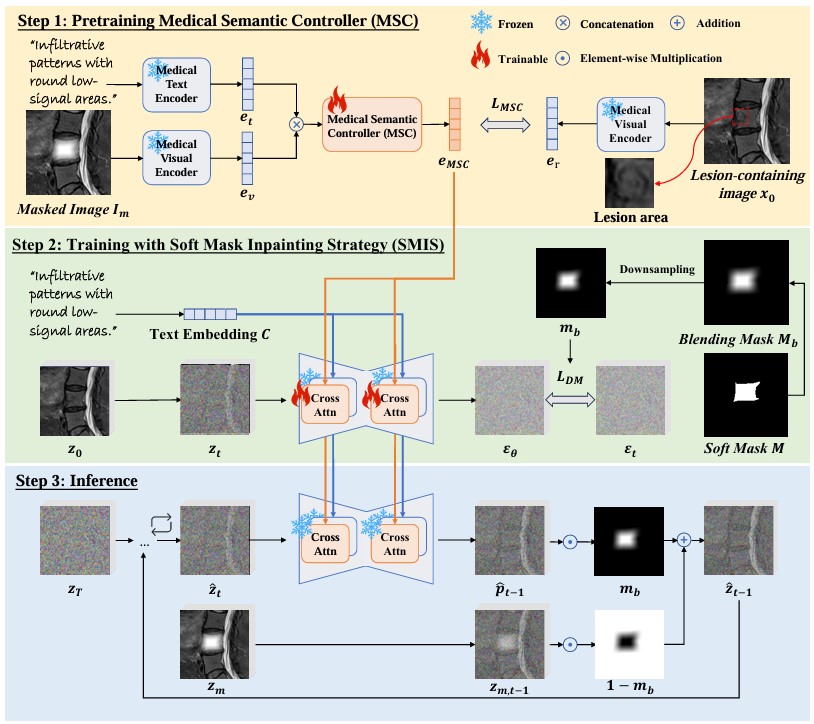
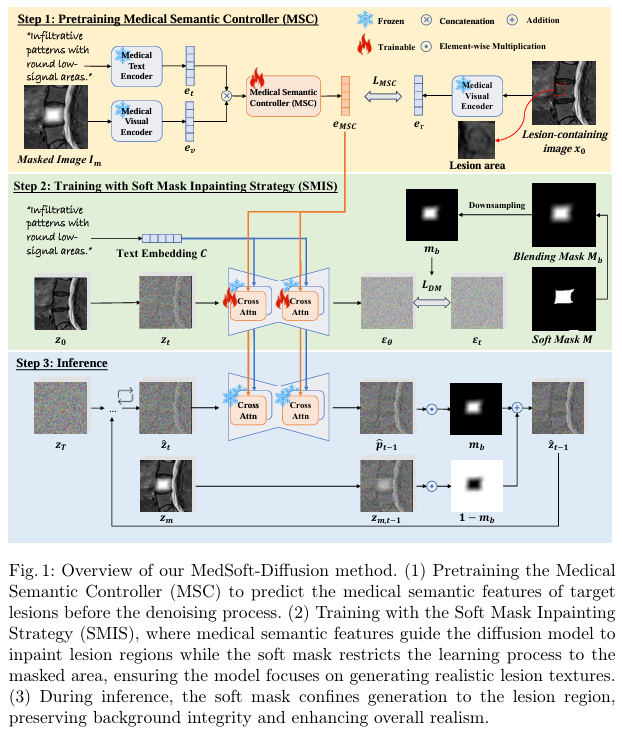
Framework
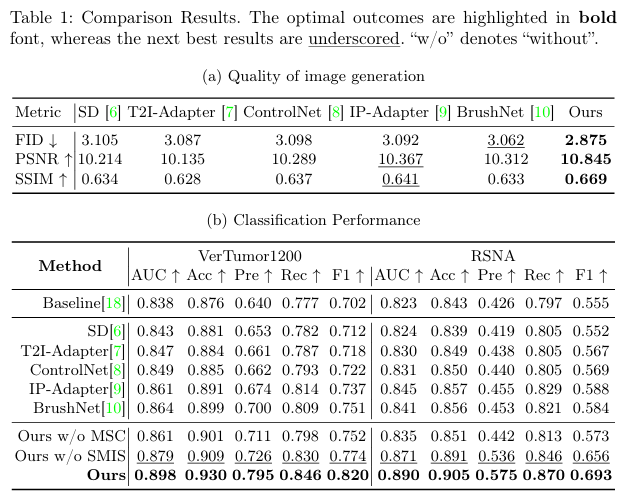
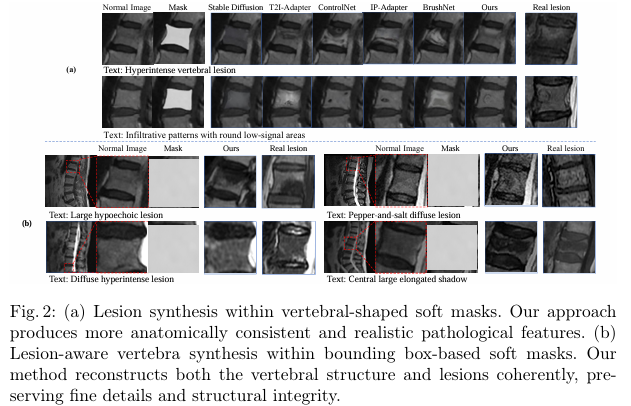
Experiment
Conclusion
In this work, we propose a novel diffusion-based framework called MedSoft Diffusion, leveraging detailed medical knowledge to generate high-quality images with specified pathological conditions that are also anatomically accurate. MSC is designed to enhance the alignment between textual prompts and lesion char acteristics, ensuring the synthesis of semantically accurate pathological images. SMIS is proposed to combine soft masks with blurring techniques to improve the realism of synthesized images. Experimental results on two vertebral disease datasets demonstrate notable improvements in both image quality and classi f ication performance using our approach. Moreover, our method can be easily extended to the synthesis of pathological images for other organs.
Acknowledgement
This work is supported by the National Key Research and De velopment Program Inter-governmental Special Project for International Science and Technology Innovation Cooperation under grant 2022YFE0112500, Foundation for Shenzhen Science and Technology Program under Grant JCYJ20240813151224032, Shenzhen Medical Research Fund under Grant B2402030 Foundation for Shenzhen Science, and Technology Program under Grant JCYJ20240813151102004.